Frequently Asked Questions: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Connecting== | |||
===How do I connect to GACRC clusters?=== | |||
Users can access GACRC clusters using secure shell (ssh) from their local machines either on-campus or off-campus. To connect via ssh, you must have an ssh software on your local machine and a connection to the UGA campus network. ssh software is included in recent releases of Unix based operating systems (including Linux and Mac OSX). If you are using a Windows computer, you can download and install PuTTY. You can find detailed instructions on how to download and install PuTTY on your Windows computer at https://wiki.gacrc.uga.edu/wiki/How_to_Install_and_Configure_PuTTY. | |||
Please note that connecting to GACRC clusters from off-campus requires connecting to the [https://eits.uga.edu/access_and_security/infosec/tools/vpn/ UGA VPN]. For more detailed information on how to connect to a specific GACRC cluster, please see the [[Connecting]] page. | |||
===I received an SSH host key error when trying to connect to a GACRC cluster. What does this mean?=== | |||
If you’ve received a warning message when attempting to connect Sapelo2 regarding the host key verification failing, this likely means you need to update your SSH known_hosts file on your local machine, by deleting the line that begins with “sapelo2.gacrc.uga.edu” (or the hostname of the GACRC machine you're trying to connect to). This can be done quickly with the following commands on Mac and Linux. This can happen as individual servers are moved into and out of our login node pool over time. | |||
Mac: | |||
<code>sed -i ' ' '/sapelo2.gacrc.uga.edu/d' ~/.ssh/known_hosts</code> | |||
Linux: | |||
<code>sed -i '/sapelo2.gacrc.uga.edu/d' ~/.ssh/known_hosts</code> | |||
After doing this, when you try to reconnect, it will ask you if you trust the computer you're connecting to. Answer affirmatively to any prompt like this, and it won't ask you that again from the computer from which you're connecting. | |||
---- | |||
[[#top|Back to Top]] | |||
==Files== | |||
===How do I copy files to/from GACRC storage?=== | |||
Users can transfer files between their local machines and GACRC storage using FTP with explicit SSL encryption, a secure copy (scp), WinSCP, FileZilla, etc. To transfer files using scp (or SSH file transfer) you must have scp (or SSH) on your local machine and a connection to the UGA campus network. An scp software is included in recent releases of Unix based operating systems (including Linux and Mac OS X). Two file transfer software that support FTP with explicit SSL encryption are the open source software FileZilla (available for Windows, Mac OS X, and Linux) and WinSCP (available for Windows machines). | |||
For more detailed information on how to copy files to/from a specific GACRC resource, please see the [[Transferring Files]] page. | |||
===Can I use text files (programs, scripts, etc) created on a Windows machine on the GACRC Unix/Linux machines?=== | |||
Text (ASCII) files created on Windows machines might have Windows newlines that are not interpreted correctly by a Unix/Linux system. However, you can convert a Windows text file to the Unix/Linux format with the dos2unix command available on the GACRC's Sapelo2 and the teaching cluster. The syntax is | |||
<code>dos2unix filename</code> | |||
where filename is the name of the ascii file (such as program.c, program.f, run.sh, input.txt, etc) created on a Windows machine. | |||
===Can I use text files (programs, scripts, etc) created on a Mac machine on the GACRC Unix/Linux machines?=== | |||
Text (ASCII) files created on Mac machines might have Mac newlines that are not interpreted correctly by a Unix/Linux system. However, you can convert a Mac text file to the Unix/Linux format with the mac2unix command available on the GACRC's Sapelo2 and the teaching cluster. The syntax is | |||
<code>mac2unix filename</code> | |||
where filename is the name of the ASCII file (such as program.c, program.f, run.sh, input.txt, etc) created on a Mac machine. | |||
===Can I leave my files in my /scratch directory?=== | |||
No, do not do this. Files not being used in /scratch will be cleaned up. Please see [https://wiki.gacrc.uga.edu/wiki/Tmp#My_data_in_.2Fscratch_disappeared._What_happened.3F the FAQ on files disappearing from /scratch] | |||
---- | |||
[[#top|Back to Top]] | |||
==Storage== | |||
===Why can't I see my lab's /project directory?=== | |||
/project directories are only accessible from the transfer nodes. Please make sure you've connected to xfer.gacrc.uga.edu (rather than the login/submit nodes) to access your lab's /project directory. Please note that /project directories are auto-mounted when you first accessed, so if you were to initially execute the command <code>ls /project</code>, you wouldn't see your lab's project directory as a subdirectory of /project, although it is there. | |||
===My data in /scratch disappeared. What happened?=== | |||
Data not being used or accessed in the /scratch file system are periodically cleaned up, as per the [https://wiki.gacrc.uga.edu/wiki/Policies#Policy_Statement_for_SCRATCH_File_System 30-day Scratch Purge Policy]. Please move your files off of /scratch when you're no longer using them. The /scratch file system is not backed up. | |||
===Is GACRC storage backed up?=== | |||
/home and /project directories are backed up, while /scratch, /work, and /lscratch are not. Please see the [https://wiki.gacrc.uga.edu/wiki/Disk_Storage#Snapshots snapshots] section of [[Disk Storage]] for more information. | |||
---- | |||
[[#top|Back to Top]] | |||
==Software== | |||
===What software is available on GACRC clusters?=== | |||
The best way to search for software on the clusters is with the <code>ml spider ''nameOfSoftware''</code> command, where ''nameOfSoftware'' is what you're searching for. You can also scroll through a full list of software modules with the <code>ml av</code> command. After entering this command, press spacebar to scroll, and q to quit. If centrally installed software has unique usage information, we document it on our [[Software]] page. In addition to software modules, we have some Singularity containers centrally installed at /apps/singularity-images on Sapelo2. | |||
===Can I install software myself on GACRC clusters?=== | |||
Yes, users can install their own software in their /home directory or their lab's /work directory. Note that this does not include installing applications from package managers such as yum or apt. Please see [[Installing Applications on Sapelo2]] for more information. | |||
===How do I access R libraries and Python modules on GACRC clusters?=== | |||
=== | ====R Libraries==== | ||
Most R libraries are added to the centrally installed R modules. Thus, in most cases, you can load the software module for the version of R that you're using and then load the desired library in your R script with <code>library(packageName)</code>. Note that we tend to not update these R libraries once they're installed, as other users could be using them. | |||
For | In some cases R libraries will have their own software module, that loads a particular version of R with it. For example, R packages that depend on the GDAL library can be found in the software module rgdal/1.5-23-foss-2019b-R-4.0.0 (for R version 4.0.0). | ||
=== | ====Python Modules==== | ||
Python modules that are not a part of the standard Python library will typically have their own software modules which also load a particular version of Python. For example, the software module TensorFlow/2.2.0-fosscuda-2019b-Python-3.7.4 would load TensorFlow version 2.2.0 and Python 3.7.4. Another example is SciPy-bundle/2020.03-fosscuda-2019b-Python-3.8.2, which loads several scientific Python packages, such as numpy and pandas, as well as Python 3.8.2. | |||
===What is Singularity?=== | |||
Please see the section on [https://wiki.gacrc.uga.edu/wiki/Software_on_Sapelo2#Singularity_Containers Singularity] in [[Software on Sapelo2]]. | |||
===How do I request an application be installed on a GACRC cluster?=== | |||
Please fill out the [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25850 software installation/update request form]. | |||
===My software requires a database, can you help?=== | |||
At this time we have very limited resources to support applications that require a database. Effectively managing a relational database is no trivial task and can require significant setup and maintenance, especially when trying to integrate one into an application on an HPC cluster. If an application allows it, it would be more efficient to use a SQLite database, which is a server-less database that creates a single database file for your application to work with, that could exist in your /scratch or /work directory while you're using it. | |||
===Can I install web services on GACRC clusters?=== | |||
Applications that are or include web services generally do not lend themselves well to HPC clusters for a variety of reasons. First of all, ports that web applications would use are not opened through the firewall on our clusters. Secondly, many web services expect to be running 24/7, which is not feasible on an HPC cluster, given that running web applications would not be acceptable on the login/submit nodes, and compute nodes are for temporary jobs, not permanent services. If there is an application you would like to use on the cluster that has a web-based component that you think may be acceptable on a GACRC cluster, please reach out to us via the [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25850 software installation/update request form] and we'll take a look at it. | |||
---- | |||
[[#top|Back to Top]] | |||
== | ==Using GACRC Clusters== | ||
===I'm brand new to high performance computing. Where do I start?=== | |||
Please see the following links to get started: | |||
* [https://kaltura.uga.edu/playlist/dedicated/176125031/1_uwkiealj/ Intro to Linux videos] | |||
* [https://kaltura.uga.edu/playlist/dedicated/176125031/1_uwkiealj/1_81u2kfi2 Intro to HPC video] | |||
* [[Best Practices on Sapelo2]] | |||
===Can I use | ===Can I use a shell other than Bash?=== | ||
When you log into a Linux machine, the environment on your terminal and the commands that you type at the prompt are defined/interpreted by a program called a shell. Examples of shells are bash, csh, ksh, tcsh, zsh. The syntax for setting environment variables and some of the functionality of your keyboard depend on the shell that you are running. For example, with bash and tcsh it is straightforward to use up arrows to recover previous commands. All users have a default shell (bash) defined at account creation time. Users who wish to have their default shell changed can request that via the [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25844 GACRC General Support] form. | |||
===Why doesn't the ls command give me colored output?=== | |||
By default <code>ls</code> does not color code its output on Sapelo2. This is because doing so required getting file metadata, which can be especially taxing on a Lustre file system (/scratch and /work) if overdone. | |||
===How do I | ===How do I use GUI applications on GACRC clusters from my Windows desktop?=== | ||
A number of software installed on GACRC | A number of software installed on GACRC clusters have X Window (GUI) front ends. Examples of such applications are Matlab, Mathematica, some text editors and debuggers, etc. In order to export such X Window applications to your Windows desktop, your desktop needs to have an X Window client (or server) running on it. A free X Window server for Microsoft Windows (10/8/7) is [http://sourceforge.net/projects/xming/ Xming]. You can download it from [http://sourceforge.net/projects/xming/ Sourceforge] and make a default installation. You will need to install the Xming server and the Xming-fonts package. Some applications also require having Xming-mesa installed. During the installation of Xming, you might want to select the option to create a desktop icon for Xming. When the installation of these two packages is complete, double click on the Xming icon to start the X Window server (a capital X will appear on your task bar). | ||
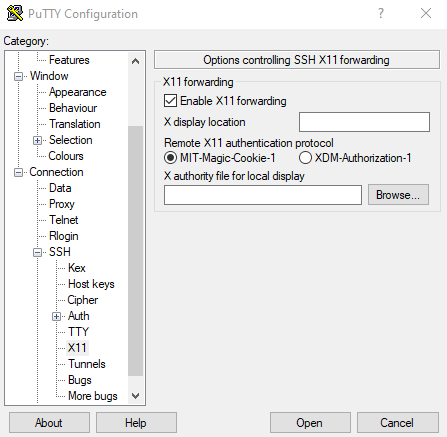
Now you need to configure your SSH client to allow tunneling of X11 connections. For example, if you use PuTTY you need to open it, expand the SSH option in the left pane, click X11 in the left pane, and check the "Enable X11 forwarding" box. | Now you need to configure your SSH client to allow tunneling of X11 connections. For example, if you use PuTTY you need to open it, expand the SSH option in the left pane, click X11 in the left pane, and check the "Enable X11 forwarding" box. | ||
| Line 69: | Line 137: | ||
Once that is done, you can SSH into your GACRC account (e.g. Sapelo2 account) and run X Window applications. The application should appear on your local Windows desktop. Each time you logout and log back into your Windows desktop, you would need to start the Xming Server manually before using PuTTY to connect to your GACRC account. | Once that is done, you can SSH into your GACRC account (e.g. Sapelo2 account) and run X Window applications. The application should appear on your local Windows desktop. Each time you logout and log back into your Windows desktop, you would need to start the Xming Server manually before using PuTTY to connect to your GACRC account. | ||
===How do I | Please note that GUI applications require a graphical interactive job session, for which more information can be found [https://wiki.gacrc.uga.edu/wiki/Running_Jobs_on_Sapelo2#How_to_run_an_interactive_job_with_Graphical_User_Interface_capabilities here] | ||
===How do I use GUI applications on GACRC clusters from my Mac?=== | |||
For Apple's OSX v10.6.3 and beyond, users have to manually install XQuartz to enable the X11 features according to [http://support.apple.com/kb/HT5293 Apple]. It is free and available at [https://www.xquartz.org/ XQuartz]. | For Apple's OSX v10.6.3 and beyond, users have to manually install XQuartz to enable the X11 features according to [http://support.apple.com/kb/HT5293 Apple]. It is free and available at [https://www.xquartz.org/ XQuartz]. | ||
| Line 89: | Line 160: | ||
After making this change on your local machine, start an XQuartz terminal and connect to sapelo2 with the '''ssh -X''' command above. | After making this change on your local machine, start an XQuartz terminal and connect to sapelo2 with the '''ssh -X''' command above. | ||
===How to | Please note that GUI applications require a graphical interactive job session, for which more information can be found [https://wiki.gacrc.uga.edu/wiki/Running_Jobs_on_Sapelo2#How_to_run_an_interactive_job_with_Graphical_User_Interface_capabilities here] | ||
===Why did I receive an email from Arbiter?=== | |||
If you've received an email from Arbiter, that means you are running a process on the login/submit nodes (ss-sub1, ss-sub2, ss-sub3, etc...) that is using a lot resources and should be run on a compute node. The login/submit nodes are only for submitting jobs to the cluster and are not for running any scientific software or scripts. If you accidentally run a process on the login/submit nodes that shouldn't be run there, Arbiter will throttle your process to preserve the integrity of the login/submit nodes for everyone else and send you an email letting you know that that happened. | |||
---- | |||
[[#top|Back to Top]] | |||
==Slurm Jobs== | |||
===How can I check on the status of my job(s)?=== | |||
* <code>squeue --me</code> - Shows the status of pending or running jobs, until a job finishes. | |||
* <code>scontrol show job ''jobid''</code> - Shows information about pending or running jobs, until very shortly after a job finishes. | |||
* <code>sacct -X -j ''jobid''</code> - Shows status/information about a job. | |||
* <code>sacct-gacrc -X -j ''jobid''</code> - <code>sacct</code> with some useful pre-formatted fields. | |||
* <code>sacct-gacrc-v ''jobid''</code> - <code>sacct-gacrc</code> displayed vertically, line by line. | |||
===I submitted my job, but I don't see anything in the output of squeue --me=== | |||
It is very likely there was a problem with your job that caused it to fail and disappear from the output of <code>squeue --me</code> before you finished typing the command. Check your Slurm job output file(s) for any errors. | |||
===Why is my job pending?=== | |||
One way that you can investigate why your job is pending is to check the rightmost column ("NODELIST/REASON") of the output of <code>squeue --me</code>. If the job is pending, rather than a list of node names on which the job is running, it will give a reason as to why the job hasn't started. These are some of the most common reasons a job may be pending: | |||
* The partition to which you have sent your job is very busy at the moment. The busier a partition is, the longer it may take for the job scheduling system to fit in your job among all the others running and waiting to run. This is also somewhat of a function of how many resources you've requested. As a general rule of thumb, the more resources requested, the longer you may have to wait for your job to start. To investigate how busy a partition is, you can use the <code>sinfo -p ''partitionName''</code> or <code>sinfo-gacrc</code> commands. | |||
* You have hit limit for the number of jobs you can have running at a time in the partition to which you've sent your job. Please see [[Job Submission partitions on Sapelo2]] for more information on how many jobs you can have running and pending at a time in a particular partition. | |||
===How do I know how much resources to request for my job?=== | |||
Please see these wiki pages to learn more about optimizing requested resources for your jobs: | |||
* [[Best Practices on Sapelo2]] | |||
* [[Job Resource Tuning]] | |||
===How much time, memory, and how many cores can I request for my jobs?=== | |||
For information on resources available in GACRC cluster partitions, please see [[Job Submission partitions on Sapelo2]] | |||
===What is an array job?=== | |||
Please see our [https://wiki.gacrc.uga.edu/wiki/Array_Jobs wiki page] on array jobs. | |||
===Can I add more time to my running job(s)?=== | |||
If your job is still running and needs more time, please reach out to us via our [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25844 general support request form], and we can add more time to it. If the job has already reached its walltime limit (and was terminated by the queueing system), it would have to be restarted. | |||
===Can I receive an email when my job starts or finishes?=== | |||
Yes. You can instruct Slurm to send you an email when your job starts or finishes with the Slurm headers --mail-user and --mail-type (defining the email address to which emails should be sent and under what conditions an email should be sent, respectively). For example: | |||
<pre class="gscript"> | |||
#!/bin/bash | |||
#SBATCH --partition=batch | |||
#SBATCH --ntasks=1 | |||
#SBATCH --mem=10gb | |||
#SBATCH --time=01:00:00 | |||
#SBATCH --mail-user=MYID@uga.edu | |||
#SBATCH --mail-type=ALL | |||
</pre> | |||
The above Slurm headers would cause an email to be sent to MYID@uga.edu when the job began, and when it finished (regardless of job success or failure). Other valid values for --mail-type include BEGIN,END,FAIL, where END would send an email when the job completes successfully, and FAIL would send an email when it finishes but fails. If you prefer to be notified when the job starts and finishes, you can just use ALL for the value of --mail-type. Note that the email address for --mail-user doesn't necessarily have to be a UGA email address, just a valid email address. | |||
---- | |||
[[#top|Back to Top]] | |||
==Training== | |||
===What training does GACRC offer?=== | |||
Every month GACRC offers Linux and Sapelo2 training for current and pending new users of Sapelo2. We also offer Python, R, and Conda training. For the current training schedule and more information, please see our [[Training]] page. | |||
===How do I sign up for GACRC training?=== | |||
To sign up for GACRC training, please fill out the [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25852 training request form]. | |||
===Is GACRC training done in person?=== | |||
No. For the foreseeable future, we will be doing our training sessions via Zoom. | |||
===Does GACRC have any training videos?=== | |||
Yes. Please see our [https://kaltura.uga.edu/channel/GACRC/176125031 Kaltura channel]. | |||
---- | |||
[[#top|Back to Top]] | |||
==Support== | |||
===How do I get GACRC support?=== | |||
The best way to get support from GACRC is to fill out the relevant form at http://help.gacrc.uga.edu. | |||
===What is the scope of GACRC support?=== | |||
We strive to provide exceptional HPC support. This is primarily focused on assistance with use of GACRC clusters. Some of the things we are able to assist our users with include but are not limited to: | |||
* Job management/troubleshooting | |||
* Data management | |||
* Software installation/troubleshooting | |||
* Script debugging/optimization | |||
* General HPC consulting | |||
* Support using Linux | |||
* Cluster account support | |||
* HPC cluster training | |||
* Programming training | |||
We cannot assist users with their actual science. This can be a gray area sometimes, but some things that are the responsibility of the researcher include but are not limited to: | |||
* Usage of scientific programs | |||
* Determining the best tool for one's research tasks | |||
* Ensuring one's input data are formatted correctly | |||
---- | |||
[[#top|Back to Top]] | |||
==Accounts== | |||
===How do I apply for accounts on GACRC clusters?=== | |||
User accounts are created as part of a "lab group" which has been registered by a Principal Investigator (PI), i.e. a UGA faculty. Once the group is registered, the PI will receive an email stating that he/she can request individual accounts for members of his/her group. For more information, please see http://gacrc.uga.edu/accounts | |||
===What do I do if I've changed lab groups or am collaborating with another lab?=== | |||
If you have switched lab groups or are collaborating with another lab group and need access to their /work and /project directories, please have the PI of your new lab group fill out the [https://uga.teamdynamix.com/TDClient/2060/Portal/Requests/ServiceDet?ID=25848 Modify/Delete Account request form]. | |||
===Will I still have access to GACRC Clusters after leaving UGA?=== | |||
As long as your MyID stays active in the UGA system and your professor/group PI wants to continue to keep you in his/her computing lab, your cluster access will be maintained by GACRC. As a student, about a year after you graduate or leave UGA, you will receive an email notifying you that your MyID account will be disabled. Faculty and staff's MyID might be disabled as soon as they leave UGA. You can find detailed info about this at | |||
https://eits.uga.edu/access_and_security/myid/myid_account_removal/. A UGA research group PI can request an 810/811 can number for a non-UGA collaborator by filling out an Affiliate form and submitting it to the UGA Card Office. The form is available at https://tate.uga.edu/wp-content/uploads/sites/4/2021/06/ugacard-affiliates-form.pdf. Once the 810/811 number is ready, the PI can contact EITS to request to keep your MyID enabled (renewed) by following the instructions at https://eits.uga.edu/access_and_security/myid/. | |||
===Will I still have access to the Teaching Cluster once the semester is over?=== | |||
Teaching cluster accounts are not long-term accounts. According to our policy, accounts created on the teaching cluster will be deleted at the end of each semester. | |||
---- | |||
[[#top|Back to Top]] | |||
==GACRC== | |||
===What compute platforms are available at GACRC?=== | |||
A list of GACRC systems, including a brief description of the compute platforms, is available at the [[Systems]] page. | |||
===How do I acknowledge the GACRC in my publication?=== | ===How do I acknowledge the GACRC in my publication?=== | ||
A sample acknowledgment statement is provided at http://gacrc.uga.edu/about/acknowledgment-statement | A sample acknowledgment statement is provided at http://gacrc.uga.edu/about/acknowledgment-statement | ||
---- | |||
[[#top|Back to Top]] | |||
Revision as of 07:33, 29 June 2021
Connecting
How do I connect to GACRC clusters?
Users can access GACRC clusters using secure shell (ssh) from their local machines either on-campus or off-campus. To connect via ssh, you must have an ssh software on your local machine and a connection to the UGA campus network. ssh software is included in recent releases of Unix based operating systems (including Linux and Mac OSX). If you are using a Windows computer, you can download and install PuTTY. You can find detailed instructions on how to download and install PuTTY on your Windows computer at https://wiki.gacrc.uga.edu/wiki/How_to_Install_and_Configure_PuTTY.
Please note that connecting to GACRC clusters from off-campus requires connecting to the UGA VPN. For more detailed information on how to connect to a specific GACRC cluster, please see the Connecting page.
I received an SSH host key error when trying to connect to a GACRC cluster. What does this mean?
If you’ve received a warning message when attempting to connect Sapelo2 regarding the host key verification failing, this likely means you need to update your SSH known_hosts file on your local machine, by deleting the line that begins with “sapelo2.gacrc.uga.edu” (or the hostname of the GACRC machine you're trying to connect to). This can be done quickly with the following commands on Mac and Linux. This can happen as individual servers are moved into and out of our login node pool over time.
Mac:
sed -i ' ' '/sapelo2.gacrc.uga.edu/d' ~/.ssh/known_hosts
Linux:
sed -i '/sapelo2.gacrc.uga.edu/d' ~/.ssh/known_hosts
After doing this, when you try to reconnect, it will ask you if you trust the computer you're connecting to. Answer affirmatively to any prompt like this, and it won't ask you that again from the computer from which you're connecting.
Files
How do I copy files to/from GACRC storage?
Users can transfer files between their local machines and GACRC storage using FTP with explicit SSL encryption, a secure copy (scp), WinSCP, FileZilla, etc. To transfer files using scp (or SSH file transfer) you must have scp (or SSH) on your local machine and a connection to the UGA campus network. An scp software is included in recent releases of Unix based operating systems (including Linux and Mac OS X). Two file transfer software that support FTP with explicit SSL encryption are the open source software FileZilla (available for Windows, Mac OS X, and Linux) and WinSCP (available for Windows machines).
For more detailed information on how to copy files to/from a specific GACRC resource, please see the Transferring Files page.
Can I use text files (programs, scripts, etc) created on a Windows machine on the GACRC Unix/Linux machines?
Text (ASCII) files created on Windows machines might have Windows newlines that are not interpreted correctly by a Unix/Linux system. However, you can convert a Windows text file to the Unix/Linux format with the dos2unix command available on the GACRC's Sapelo2 and the teaching cluster. The syntax is
dos2unix filename
where filename is the name of the ascii file (such as program.c, program.f, run.sh, input.txt, etc) created on a Windows machine.
Can I use text files (programs, scripts, etc) created on a Mac machine on the GACRC Unix/Linux machines?
Text (ASCII) files created on Mac machines might have Mac newlines that are not interpreted correctly by a Unix/Linux system. However, you can convert a Mac text file to the Unix/Linux format with the mac2unix command available on the GACRC's Sapelo2 and the teaching cluster. The syntax is
mac2unix filename
where filename is the name of the ASCII file (such as program.c, program.f, run.sh, input.txt, etc) created on a Mac machine.
Can I leave my files in my /scratch directory?
No, do not do this. Files not being used in /scratch will be cleaned up. Please see the FAQ on files disappearing from /scratch
Storage
Why can't I see my lab's /project directory?
/project directories are only accessible from the transfer nodes. Please make sure you've connected to xfer.gacrc.uga.edu (rather than the login/submit nodes) to access your lab's /project directory. Please note that /project directories are auto-mounted when you first accessed, so if you were to initially execute the command ls /project, you wouldn't see your lab's project directory as a subdirectory of /project, although it is there.
My data in /scratch disappeared. What happened?
Data not being used or accessed in the /scratch file system are periodically cleaned up, as per the 30-day Scratch Purge Policy. Please move your files off of /scratch when you're no longer using them. The /scratch file system is not backed up.
Is GACRC storage backed up?
/home and /project directories are backed up, while /scratch, /work, and /lscratch are not. Please see the snapshots section of Disk Storage for more information.
Software
What software is available on GACRC clusters?
The best way to search for software on the clusters is with the ml spider nameOfSoftware command, where nameOfSoftware is what you're searching for. You can also scroll through a full list of software modules with the ml av command. After entering this command, press spacebar to scroll, and q to quit. If centrally installed software has unique usage information, we document it on our Software page. In addition to software modules, we have some Singularity containers centrally installed at /apps/singularity-images on Sapelo2.
Can I install software myself on GACRC clusters?
Yes, users can install their own software in their /home directory or their lab's /work directory. Note that this does not include installing applications from package managers such as yum or apt. Please see Installing Applications on Sapelo2 for more information.
How do I access R libraries and Python modules on GACRC clusters?
R Libraries
Most R libraries are added to the centrally installed R modules. Thus, in most cases, you can load the software module for the version of R that you're using and then load the desired library in your R script with library(packageName). Note that we tend to not update these R libraries once they're installed, as other users could be using them.
In some cases R libraries will have their own software module, that loads a particular version of R with it. For example, R packages that depend on the GDAL library can be found in the software module rgdal/1.5-23-foss-2019b-R-4.0.0 (for R version 4.0.0).
Python Modules
Python modules that are not a part of the standard Python library will typically have their own software modules which also load a particular version of Python. For example, the software module TensorFlow/2.2.0-fosscuda-2019b-Python-3.7.4 would load TensorFlow version 2.2.0 and Python 3.7.4. Another example is SciPy-bundle/2020.03-fosscuda-2019b-Python-3.8.2, which loads several scientific Python packages, such as numpy and pandas, as well as Python 3.8.2.
What is Singularity?
Please see the section on Singularity in Software on Sapelo2.
How do I request an application be installed on a GACRC cluster?
Please fill out the software installation/update request form.
My software requires a database, can you help?
At this time we have very limited resources to support applications that require a database. Effectively managing a relational database is no trivial task and can require significant setup and maintenance, especially when trying to integrate one into an application on an HPC cluster. If an application allows it, it would be more efficient to use a SQLite database, which is a server-less database that creates a single database file for your application to work with, that could exist in your /scratch or /work directory while you're using it.
Can I install web services on GACRC clusters?
Applications that are or include web services generally do not lend themselves well to HPC clusters for a variety of reasons. First of all, ports that web applications would use are not opened through the firewall on our clusters. Secondly, many web services expect to be running 24/7, which is not feasible on an HPC cluster, given that running web applications would not be acceptable on the login/submit nodes, and compute nodes are for temporary jobs, not permanent services. If there is an application you would like to use on the cluster that has a web-based component that you think may be acceptable on a GACRC cluster, please reach out to us via the software installation/update request form and we'll take a look at it.
Using GACRC Clusters
I'm brand new to high performance computing. Where do I start?
Please see the following links to get started:
Can I use a shell other than Bash?
When you log into a Linux machine, the environment on your terminal and the commands that you type at the prompt are defined/interpreted by a program called a shell. Examples of shells are bash, csh, ksh, tcsh, zsh. The syntax for setting environment variables and some of the functionality of your keyboard depend on the shell that you are running. For example, with bash and tcsh it is straightforward to use up arrows to recover previous commands. All users have a default shell (bash) defined at account creation time. Users who wish to have their default shell changed can request that via the GACRC General Support form.
Why doesn't the ls command give me colored output?
By default ls does not color code its output on Sapelo2. This is because doing so required getting file metadata, which can be especially taxing on a Lustre file system (/scratch and /work) if overdone.
How do I use GUI applications on GACRC clusters from my Windows desktop?
A number of software installed on GACRC clusters have X Window (GUI) front ends. Examples of such applications are Matlab, Mathematica, some text editors and debuggers, etc. In order to export such X Window applications to your Windows desktop, your desktop needs to have an X Window client (or server) running on it. A free X Window server for Microsoft Windows (10/8/7) is Xming. You can download it from Sourceforge and make a default installation. You will need to install the Xming server and the Xming-fonts package. Some applications also require having Xming-mesa installed. During the installation of Xming, you might want to select the option to create a desktop icon for Xming. When the installation of these two packages is complete, double click on the Xming icon to start the X Window server (a capital X will appear on your task bar).
Now you need to configure your SSH client to allow tunneling of X11 connections. For example, if you use PuTTY you need to open it, expand the SSH option in the left pane, click X11 in the left pane, and check the "Enable X11 forwarding" box.
Once that is done, you can SSH into your GACRC account (e.g. Sapelo2 account) and run X Window applications. The application should appear on your local Windows desktop. Each time you logout and log back into your Windows desktop, you would need to start the Xming Server manually before using PuTTY to connect to your GACRC account.
Please note that GUI applications require a graphical interactive job session, for which more information can be found here
How do I use GUI applications on GACRC clusters from my Mac?
For Apple's OSX v10.6.3 and beyond, users have to manually install XQuartz to enable the X11 features according to Apple. It is free and available at XQuartz.
Then connect to Sapelo as:
ssh -X myid@sapelo2.gacrc.uga.edu
Please check where your local machine has xauth installed, e.g. is it in /opt/X11/bin/xauth or somewhere else? Then edit the ~/.ssh/config file on your local machine (not on Sapelo2) to add the location of xauth, e.g. add
Host * XAuthLocation /opt/X11/bin/xauth
if that is the path of xauth on your machine. If ~/.ssh/config does not exist, create this file and put the lines above in this file.
After making this change on your local machine, start an XQuartz terminal and connect to sapelo2 with the ssh -X command above.
Please note that GUI applications require a graphical interactive job session, for which more information can be found here
Why did I receive an email from Arbiter?
If you've received an email from Arbiter, that means you are running a process on the login/submit nodes (ss-sub1, ss-sub2, ss-sub3, etc...) that is using a lot resources and should be run on a compute node. The login/submit nodes are only for submitting jobs to the cluster and are not for running any scientific software or scripts. If you accidentally run a process on the login/submit nodes that shouldn't be run there, Arbiter will throttle your process to preserve the integrity of the login/submit nodes for everyone else and send you an email letting you know that that happened.
Slurm Jobs
How can I check on the status of my job(s)?
squeue --me- Shows the status of pending or running jobs, until a job finishes.scontrol show job jobid- Shows information about pending or running jobs, until very shortly after a job finishes.sacct -X -j jobid- Shows status/information about a job.sacct-gacrc -X -j jobid-sacctwith some useful pre-formatted fields.sacct-gacrc-v jobid-sacct-gacrcdisplayed vertically, line by line.
I submitted my job, but I don't see anything in the output of squeue --me
It is very likely there was a problem with your job that caused it to fail and disappear from the output of squeue --me before you finished typing the command. Check your Slurm job output file(s) for any errors.
Why is my job pending?
One way that you can investigate why your job is pending is to check the rightmost column ("NODELIST/REASON") of the output of squeue --me. If the job is pending, rather than a list of node names on which the job is running, it will give a reason as to why the job hasn't started. These are some of the most common reasons a job may be pending:
- The partition to which you have sent your job is very busy at the moment. The busier a partition is, the longer it may take for the job scheduling system to fit in your job among all the others running and waiting to run. This is also somewhat of a function of how many resources you've requested. As a general rule of thumb, the more resources requested, the longer you may have to wait for your job to start. To investigate how busy a partition is, you can use the
sinfo -p partitionNameorsinfo-gacrccommands. - You have hit limit for the number of jobs you can have running at a time in the partition to which you've sent your job. Please see Job Submission partitions on Sapelo2 for more information on how many jobs you can have running and pending at a time in a particular partition.
How do I know how much resources to request for my job?
Please see these wiki pages to learn more about optimizing requested resources for your jobs:
How much time, memory, and how many cores can I request for my jobs?
For information on resources available in GACRC cluster partitions, please see Job Submission partitions on Sapelo2
What is an array job?
Please see our wiki page on array jobs.
Can I add more time to my running job(s)?
If your job is still running and needs more time, please reach out to us via our general support request form, and we can add more time to it. If the job has already reached its walltime limit (and was terminated by the queueing system), it would have to be restarted.
Can I receive an email when my job starts or finishes?
Yes. You can instruct Slurm to send you an email when your job starts or finishes with the Slurm headers --mail-user and --mail-type (defining the email address to which emails should be sent and under what conditions an email should be sent, respectively). For example:
#!/bin/bash #SBATCH --partition=batch #SBATCH --ntasks=1 #SBATCH --mem=10gb #SBATCH --time=01:00:00 #SBATCH --mail-user=MYID@uga.edu #SBATCH --mail-type=ALL
The above Slurm headers would cause an email to be sent to MYID@uga.edu when the job began, and when it finished (regardless of job success or failure). Other valid values for --mail-type include BEGIN,END,FAIL, where END would send an email when the job completes successfully, and FAIL would send an email when it finishes but fails. If you prefer to be notified when the job starts and finishes, you can just use ALL for the value of --mail-type. Note that the email address for --mail-user doesn't necessarily have to be a UGA email address, just a valid email address.
Training
What training does GACRC offer?
Every month GACRC offers Linux and Sapelo2 training for current and pending new users of Sapelo2. We also offer Python, R, and Conda training. For the current training schedule and more information, please see our Training page.
How do I sign up for GACRC training?
To sign up for GACRC training, please fill out the training request form.
Is GACRC training done in person?
No. For the foreseeable future, we will be doing our training sessions via Zoom.
Does GACRC have any training videos?
Yes. Please see our Kaltura channel.
Support
How do I get GACRC support?
The best way to get support from GACRC is to fill out the relevant form at http://help.gacrc.uga.edu.
What is the scope of GACRC support?
We strive to provide exceptional HPC support. This is primarily focused on assistance with use of GACRC clusters. Some of the things we are able to assist our users with include but are not limited to:
- Job management/troubleshooting
- Data management
- Software installation/troubleshooting
- Script debugging/optimization
- General HPC consulting
- Support using Linux
- Cluster account support
- HPC cluster training
- Programming training
We cannot assist users with their actual science. This can be a gray area sometimes, but some things that are the responsibility of the researcher include but are not limited to:
- Usage of scientific programs
- Determining the best tool for one's research tasks
- Ensuring one's input data are formatted correctly
Accounts
How do I apply for accounts on GACRC clusters?
User accounts are created as part of a "lab group" which has been registered by a Principal Investigator (PI), i.e. a UGA faculty. Once the group is registered, the PI will receive an email stating that he/she can request individual accounts for members of his/her group. For more information, please see http://gacrc.uga.edu/accounts
What do I do if I've changed lab groups or am collaborating with another lab?
If you have switched lab groups or are collaborating with another lab group and need access to their /work and /project directories, please have the PI of your new lab group fill out the Modify/Delete Account request form.
Will I still have access to GACRC Clusters after leaving UGA?
As long as your MyID stays active in the UGA system and your professor/group PI wants to continue to keep you in his/her computing lab, your cluster access will be maintained by GACRC. As a student, about a year after you graduate or leave UGA, you will receive an email notifying you that your MyID account will be disabled. Faculty and staff's MyID might be disabled as soon as they leave UGA. You can find detailed info about this at https://eits.uga.edu/access_and_security/myid/myid_account_removal/. A UGA research group PI can request an 810/811 can number for a non-UGA collaborator by filling out an Affiliate form and submitting it to the UGA Card Office. The form is available at https://tate.uga.edu/wp-content/uploads/sites/4/2021/06/ugacard-affiliates-form.pdf. Once the 810/811 number is ready, the PI can contact EITS to request to keep your MyID enabled (renewed) by following the instructions at https://eits.uga.edu/access_and_security/myid/.
Will I still have access to the Teaching Cluster once the semester is over?
Teaching cluster accounts are not long-term accounts. According to our policy, accounts created on the teaching cluster will be deleted at the end of each semester.
GACRC
What compute platforms are available at GACRC?
A list of GACRC systems, including a brief description of the compute platforms, is available at the Systems page.
How do I acknowledge the GACRC in my publication?
A sample acknowledgment statement is provided at http://gacrc.uga.edu/about/acknowledgment-statement