SRAToolKit-Sapelo2: Difference between revisions
No edit summary |
|||
| (13 intermediate revisions by 5 users not shown) | |||
| Line 5: | Line 5: | ||
Sapelo2 | Sapelo2 | ||
=== Version === | === Version === | ||
3.0.1, 3.0.3 | |||
===Author / Distributor=== | ===Author / Distributor=== | ||
Please see https://github.com/ncbi/sra-tools | Please see https://github.com/ncbi/sra-tools | ||
===Description=== | ===Description=== | ||
The SRA Toolkit from NCBI is a collection of tools and libraries for using data in the INSDC Sequence Read Archives. The Sequence Read Archives (SRA) store raw sequence data from "next-generation" sequencing technologies including Illumina, 454, IonTorrent, Complete Genomics, PacBio and OxfordNanopores. In addition to raw sequence data, SRA now stores alignment information in the form of read placements on a reference sequence. The SRA-Toolkit includes the following tools: | The SRA Toolkit from NCBI is a collection of tools and libraries for using data in the INSDC Sequence Read Archives. The Sequence Read Archives (SRA) store raw sequence data from "next-generation" sequencing technologies including Illumina, 454, IonTorrent, Complete Genomics, PacBio and OxfordNanopores. In addition to raw sequence data, SRA now stores alignment information in the form of read placements on a reference sequence. The SRA-Toolkit includes the following tools: | ||
{| class="wikitable" | |||
|+ | |||
!Command | |||
!Description | |||
|- | |||
|<code>fastq-dump</code> | |||
|Convert SRA data into fastq format | |||
|- | |||
|<code>prefetch</code> | |||
|Allows command-line downloading of SRA, dbGaP, and ADSP data | |||
|- | |||
|<code>sam-dump</code> | |||
|Convert SRA data to sam format | |||
|- | |||
|<code>sra-pileup</code> | |||
|Generate pileup statistics on aligned SRA data | |||
|- | |||
|<code>vdb-config</code> | |||
|Display and modify VDB configuration information | |||
|- | |||
|<code>vdb-decrypt</code> | |||
|Decrypt non-SRA dbGaP data ("phenotype data") | |||
|- | |||
|<code>abi-dump</code> | |||
|Convert SRA data into ABI format (csfasta / qual) | |||
|- | |||
|<code>illumina-dump</code> | |||
|Convert SRA data into Illumina native formats (qseq, etc.) | |||
|- | |||
|<code>sff-dump</code> | |||
|Convert SRA data to sff format | |||
|- | |||
|<code>sra-stat</code> | |||
|Generate statistics about SRA data (quality distribution, etc.) | |||
|- | |||
|<code>vdb-dump</code> | |||
|Output the native VDB format of SRA data. | |||
|- | |||
|<code>vdb-encrypt</code> | |||
|Encrypt non-SRA dbGaP data ("phenotype data") | |||
|- | |||
|<code>vdb-validate</code> | |||
|Validate the integrity of downloaded SRA data | |||
|} | |||
=== Downloading SRA Data === | |||
The last version of this application is at /apps/eb/SRA-Toolkit/3.0.3-gompi-2022a | |||
To use this version, please load the module with | |||
<pre class="gscript"> | |||
ml SRA-Toolkit/3.0.3-gompi-2022a | |||
</pre> | |||
You can download SRA data to local directory with the prefetch tool. This program downloads Runs (sequence files in the compressed SRA format) and all additional data necessary to convert the Run from the SRA format to a more commonly used format. You can search for a dataset using the search bar at the top of the SRA homepage. https://www.ncbi.nlm.nih.gov/sra Once you find a dataset you would like to download, search for the "Run number" in the table towards the bottom of the webpage for that dataset. Then create the folder where prefetch will deposit your files. This needs to be an empty folder. It is recommended that you download data into your scratch directory since there is plenty of space there. | |||
Next, run the command: | |||
vdb- | <pre>vdb-config --interactive</pre> | ||
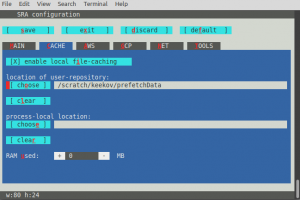
This will open a screen where you operate the buttons by pressing the letter highlighted in red, or by pressing the tab-key until the wanted button is reached and then pressing the space- or the enter-key. Make sure there is an X by the "'''Enable Remote Access'''" option on the MAIN tab, and X by the "'''enable local file-caching'''" option in the CACHE tab. Then set the "location of user-repository" to the empty folder you created. In the following image the data will be downloaded to /scratch/keekov/prefetchData. These settings are stored in a file called '''user-settings.mkfg''' in a hidden directory called .ncbi in the top level of your home directory. | |||
[[File:Sratools.png|thumb]] | |||
Then press "s" or navigate to the save button and press enter to save. Then press "x" or navigate to the exit button and press enter to exit. Now you can start the data download by running the command prefetch followed by the run number. For example the following downloads the dataset SRR390728. | |||
<pre>prefetch SRR390728</pre> | |||
For more information about the prefetch command refer to the [https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=toolkit_doc&f=prefetch documentation] | |||
=== Running Program === | === Running Program === | ||
You can now run other SRA-Tools on the SRA data you've downloaded. | |||
Also refer to [[Running Jobs on Sapelo2]] | Also refer to [[Running Jobs on Sapelo2]] | ||
For more information on Environment Modules on Sapelo please see the [[Lmod]] page. | For more information on Environment Modules on Sapelo please see the [[Lmod]] page. | ||
Here is an example of a shell script, sub.sh, to run on the batch queue: | |||
<div class="gscript2"> | |||
<nowiki>#</nowiki>!/bin/bash<br> | |||
<nowiki>#</nowiki>SBATCH --job-name=SRA-ToolsExample<br> | |||
<nowiki>#</nowiki>SBATCH --partition=batch<br> | |||
<nowiki>#</nowiki>SBATCH --mail-type=ALL<br> | |||
<nowiki>#</nowiki>SBATCH --mail-user=<u>username@uga.edu</u><br> | |||
<nowiki>#</nowiki>SBATCH --ntasks=<u>1</u><br> | |||
<nowiki>#</nowiki>SBATCH --mem=<u>10gb</u><br> | |||
<nowiki>#</nowiki>SBATCH --time=<u>08:00:00</u><br> | |||
<nowiki>#</nowiki>SBATCH --output=fastq-dump.%j.out<br> | |||
<nowiki>#</nowiki>SBATCH --error=fastq-dump.%j.err<br> | |||
cd $SLURM_SUBMIT_DIR<br> | |||
ml SRA-Toolkit/3.0.3-gompi-2022a<br> | |||
fastq-dump -X 5 -Z /scratch/keekov/SRR390728.sra #You should use the directory you selected in the vdb-config step above<br> | |||
</div> | |||
=== Documentation === | === Documentation === | ||
Latest revision as of 14:53, 25 June 2024
Category
BioInformatics
Program On
Sapelo2
Version
3.0.1, 3.0.3
Author / Distributor
Please see https://github.com/ncbi/sra-tools
Description
The SRA Toolkit from NCBI is a collection of tools and libraries for using data in the INSDC Sequence Read Archives. The Sequence Read Archives (SRA) store raw sequence data from "next-generation" sequencing technologies including Illumina, 454, IonTorrent, Complete Genomics, PacBio and OxfordNanopores. In addition to raw sequence data, SRA now stores alignment information in the form of read placements on a reference sequence. The SRA-Toolkit includes the following tools:
| Command | Description |
|---|---|
fastq-dump
|
Convert SRA data into fastq format |
prefetch
|
Allows command-line downloading of SRA, dbGaP, and ADSP data |
sam-dump
|
Convert SRA data to sam format |
sra-pileup
|
Generate pileup statistics on aligned SRA data |
vdb-config
|
Display and modify VDB configuration information |
vdb-decrypt
|
Decrypt non-SRA dbGaP data ("phenotype data") |
abi-dump
|
Convert SRA data into ABI format (csfasta / qual) |
illumina-dump
|
Convert SRA data into Illumina native formats (qseq, etc.) |
sff-dump
|
Convert SRA data to sff format |
sra-stat
|
Generate statistics about SRA data (quality distribution, etc.) |
vdb-dump
|
Output the native VDB format of SRA data. |
vdb-encrypt
|
Encrypt non-SRA dbGaP data ("phenotype data") |
vdb-validate
|
Validate the integrity of downloaded SRA data |
Downloading SRA Data
The last version of this application is at /apps/eb/SRA-Toolkit/3.0.3-gompi-2022a
To use this version, please load the module with
ml SRA-Toolkit/3.0.3-gompi-2022a
You can download SRA data to local directory with the prefetch tool. This program downloads Runs (sequence files in the compressed SRA format) and all additional data necessary to convert the Run from the SRA format to a more commonly used format. You can search for a dataset using the search bar at the top of the SRA homepage. https://www.ncbi.nlm.nih.gov/sra Once you find a dataset you would like to download, search for the "Run number" in the table towards the bottom of the webpage for that dataset. Then create the folder where prefetch will deposit your files. This needs to be an empty folder. It is recommended that you download data into your scratch directory since there is plenty of space there.
Next, run the command:
vdb-config --interactive
This will open a screen where you operate the buttons by pressing the letter highlighted in red, or by pressing the tab-key until the wanted button is reached and then pressing the space- or the enter-key. Make sure there is an X by the "Enable Remote Access" option on the MAIN tab, and X by the "enable local file-caching" option in the CACHE tab. Then set the "location of user-repository" to the empty folder you created. In the following image the data will be downloaded to /scratch/keekov/prefetchData. These settings are stored in a file called user-settings.mkfg in a hidden directory called .ncbi in the top level of your home directory.
Then press "s" or navigate to the save button and press enter to save. Then press "x" or navigate to the exit button and press enter to exit. Now you can start the data download by running the command prefetch followed by the run number. For example the following downloads the dataset SRR390728.
prefetch SRR390728
For more information about the prefetch command refer to the documentation
Running Program
You can now run other SRA-Tools on the SRA data you've downloaded. Also refer to Running Jobs on Sapelo2
For more information on Environment Modules on Sapelo please see the Lmod page.
Here is an example of a shell script, sub.sh, to run on the batch queue:
#!/bin/bash
#SBATCH --job-name=SRA-ToolsExample
#SBATCH --partition=batch
#SBATCH --mail-type=ALL
#SBATCH --mail-user=username@uga.edu
#SBATCH --ntasks=1
#SBATCH --mem=10gb
#SBATCH --time=08:00:00
#SBATCH --output=fastq-dump.%j.out
#SBATCH --error=fastq-dump.%j.err
cd $SLURM_SUBMIT_DIR
ml SRA-Toolkit/3.0.3-gompi-2022a
fastq-dump -X 5 -Z /scratch/keekov/SRR390728.sra #You should use the directory you selected in the vdb-config step above
Documentation
Please see https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=toolkit_doc for the documentation of each tool.
Installation
System
64-bit Linux